On June 13, 2025, we took part in the PINKCC Challenge final at ICM in Montpellier, organized with SIRIC Montpellier. The goal of the challenge was to segment tumors and metastases in CT scans from ovarian cancer patients. We used nnUNetv1 for our solution, training three different setups with 5-fold cross-validation, then combined the results using ensembling. The idea was to keep things simple and solid — and it worked out well.
nnUNet: The Go-To Framework for Medical Image Segmentation
The nnUNet framework has become a standard in medical image segmentation. Recognized for its state-of-the-art performance, it has consistently ranked at the top of various segmentation benchmarks and challenges (
Isensee et al., 2019). What makes nnUNet stand out is its simplicity — it doesn’t rely on complex or custom architectures, but instead focuses on strong training protocols, automated configuration, and sensible defaults.
We followed this philosophy in our participation in the PINKCC Challenge, which focused on segmenting primary tumors and metastases in CT scans of ovarian cancer patients. This is a difficult task due to the wide variability in lesion size, location, and imaging quality.
For our solution, we used nnUNetv1, training three configurations using 5-fold cross-validation, and combined their predictions through ensembling.
✅ ResNet-based multiclass model: segments both primary tumors and metastases.
✅ Multiclass model with isotropic spacing: improves handling of 3D volume consistency.
✅ UNet model for metastasis-only segmentation: focused specifically on small lesion sensitivity.
The full pipeline is illustrated below:
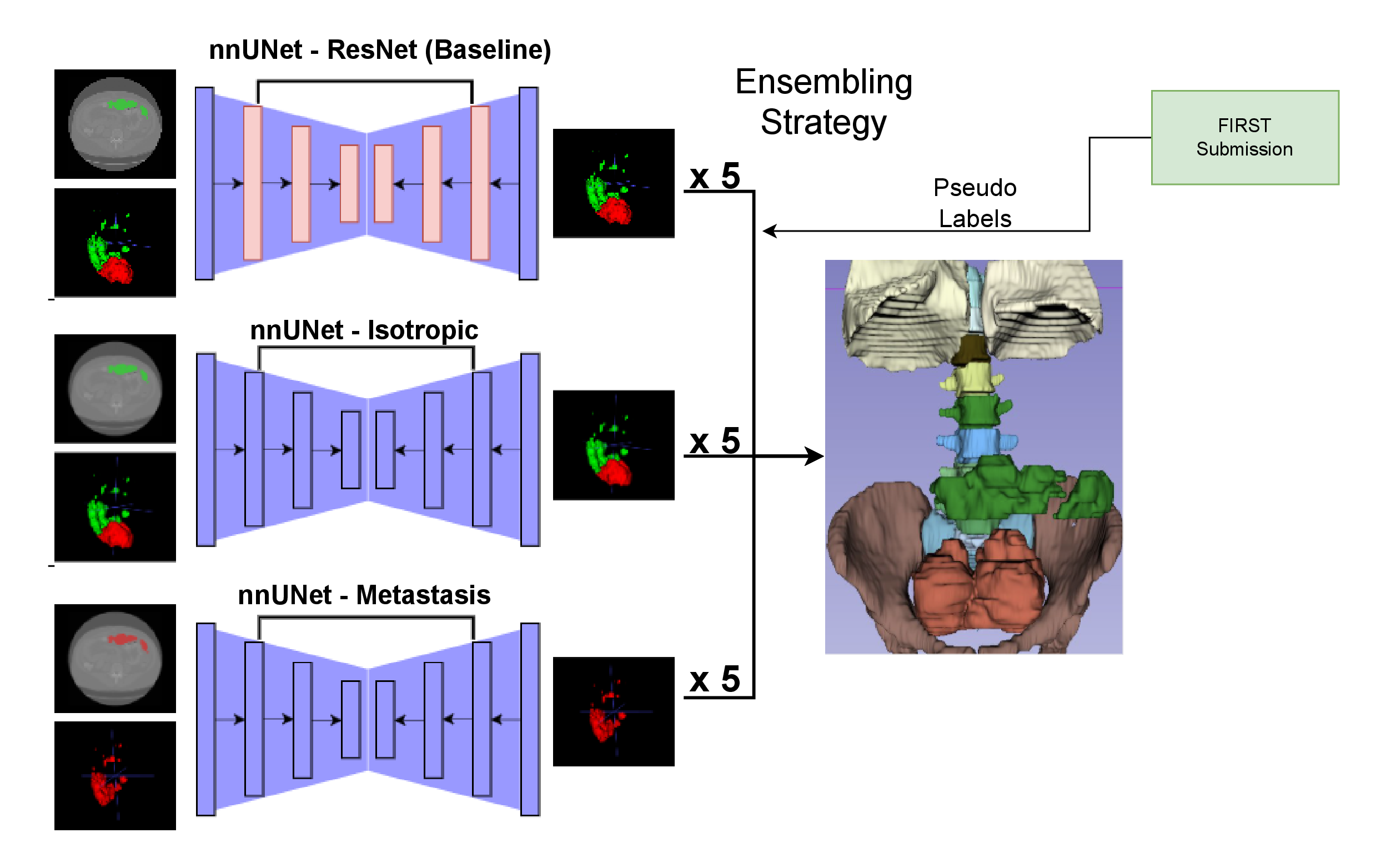
We started with careful data quality control, checking image orientation, annotation consistency, and label distribution. Each of the three nnUNet variants had its own tailored preprocessing setup. We trained five models per configuration (5-fold), and ensembled their outputs to increase robustness. As a final step, we used TotalSegmentator to post-process and refine the results for cleaner, more accurate segmentations.
The code for our pipeline is publicly available at:
https://github.com/Andrade-Miranda/PINKCC_CERIS

Team CERIS in action during the PINKCC Challenge — collaborative effort, focus, and some coffee-powered segmentation breakthroughs.


